Photo from wikipedia
Protein engineering has an array of uses: whether you are studying a disease mutation, removing undesirable sequences, adding stabilizing mutations for structural purposes, or simply dissecting protein function. Protein engineering… Click to show full abstract
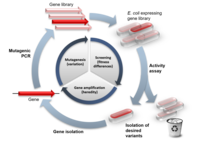
Protein engineering has an array of uses: whether you are studying a disease mutation, removing undesirable sequences, adding stabilizing mutations for structural purposes, or simply dissecting protein function. Protein engineering is almost exclusively performed using site-directed mutagenesis (SDM) as this provides targeted modification of specific amino acids, as well as the option of rewriting the native sequence to include or exclude certain regions. Despite its widespread use, SDM has often proved to be a bottleneck, requiring precision manipulation on a sample-by-sample basis to make it work. When dealing with large volumes of samples it is not possible to use such a low-throughput approach. Here we describe a high-throughput (HTP) method for SDM, optimized and used by the Structural Genomics Consortium (SGC) to complement structural studies.
Journal Title: Methods in molecular biology
Year Published: 2019
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!