Photo from wikipedia
Antibiotic resistance of Salmonella species is well reported. Ciprofloxacin is the frontline antibiotic for salmonellosis. The repeated exposure to ciprofloxacin leads to resistant strains. After 20 cycles of antibiotic exposure,… Click to show full abstract
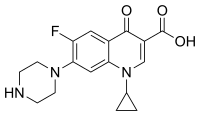
Antibiotic resistance of Salmonella species is well reported. Ciprofloxacin is the frontline antibiotic for salmonellosis. The repeated exposure to ciprofloxacin leads to resistant strains. After 20 cycles of antibiotic exposure, resistant bacterial clones were evaluated. The colony size of the mutants was small and had an extended lag phase compared to parent strain. The whole genome sequencing showed 40,513 mutations across the genome. Small percentage (5.2%) of mutations was non-synonymous. Four-fold more transitions were observed than transversions. Ratio of < 1 transition vs transversion showed a positive selection for antibiotic resistant trait. Mutation distribution across the genome was uniform. The native plasmid was an exception and 2 mutations were observed on 90 kb plasmid. The important genes like dnaE, gyrA, iroC, metH and rpoB involved in antibiotic resistance had point mutations. The genome analysis revealed most of the metabolic pathways were affected.
Journal Title: Archives of microbiology
Year Published: 2021
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!