Photo from wikipedia
A total of 33,206 wild boars (Sus scrofa) hunted throughout the province of Girona (northeastern Spain) were submitted to a game processing plant during four hunting seasons (2014–2015 to 2017–2018).… Click to show full abstract
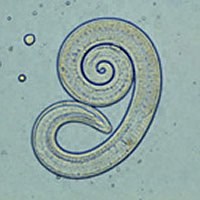
A total of 33,206 wild boars (Sus scrofa) hunted throughout the province of Girona (northeastern Spain) were submitted to a game processing plant during four hunting seasons (2014–2015 to 2017–2018). This province is divided into seven districts with different habitat types for wild boar populations. Muscle samples from each wild boar were tested for Trichinella spp. infection by artificial digestion following Regulations 2075/2005 and 2015/1375 of the European Union. Larvae from positive muscle samples were characterized at the species level in the National Reference Laboratory for Trichinella in Spain using a multiplex PCR analysis. Trichinella larvae were found in a total 112 wild boars (0.34%; 95% IC 0.27–0.39%). Results indicated an annual ascending trend in the percentage of infected wild boars; however, this trend was small, as shown by the low values of both Kendall’s tau (0.328; p=0.012) and the linear fit slope (b=8.864E-5). The prevalence of Trichinella spp. per district and hunting season was positively correlated with hunting yield (Spearman’s rank correlation rs = 0.440; p = 0.024). Spring prevalence (0%) was significantly lower than in other seasons (0.29–0.33%), although this effect could be explained by the scarcity of captures during the months of April, May, and June. The finding of one positive animal weighing only 7.5 kg indicates that piglets can be infected by Trichinella at an early age, but a significantly higher proportion of positives was found among adult wild boars exceeding 50 kg. The molecular identification of 112 Trichinella isolates revealed the predominance of Trichinella spiralis (n = 109) over Trichinella britovi (n = 3). These findings indicate that Trichinella infection in wild boar remains a public health problem in northeastern Spain.
Journal Title: European Journal of Wildlife Research
Year Published: 2021
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!