Photo from wikipedia
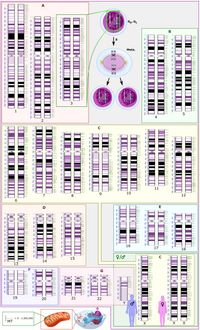
Chaling common wild rice (Chaling CWR, Oryza rufipogon Griff.) is a kind of germplasm resource which possesses abundant genetic diversity and plays a pivotal role in improving cultivated rice. In this… Click to show full abstract
Chaling common wild rice (Chaling CWR, Oryza rufipogon Griff.) is a kind of germplasm resource which possesses abundant genetic diversity and plays a pivotal role in improving cultivated rice. In this research, we used next-generation sequencing technology to reveal the genome sequence of Chaling CWR and investigate its elite genes. The results showed that the average coverage was 93.99%. 1,743,368 single-nucleotide polymorphism (SNP) and 387,381 insertion and deletion (InDel) variants have been detected in Chaling CWR compared with the Nipponbare genome. The variant density was found to be highest in chromosome 10 and lowest in chromosome 5, with an average density of 467.08 SNPs/100 kb and 109.59 InDels/100 kb across the whole genome. 109,684 non-synonymous SNPs were identified in 14,315 genes, and 26,051 InDels were detected in the coding regions. We further classified these genes based on the protein domains that were involved in host defense mechanisms and in the analysis of 37 NBS-LRRs, 71 LRRs, 136 NB-ARCs, 119 PKs, 19 WRKY transcription factors, and 25 resistant protein genes. Among the putative R-genes, Pish, Pita, Pi25, Pi9, Pi21, Pid2, Pikm, and Xa21 were further identified, as exhibiting high rates of polymorphism. Results from this study provide a foundation for future basic research and marker-assisted breeding of rice resistance.
Journal Title: Plant Molecular Biology Reporter
Year Published: 2020
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!