Photo from wikipedia
This study aimed to investigate sites for colonization and molecular epidemiology of antimicrobial-resistant Pseudomonas aeruginosa in a veterinary teaching hospital. Bacterial specimens from surface and liquid samples (n = 165) located in… Click to show full abstract
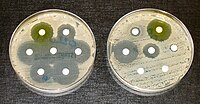
This study aimed to investigate sites for colonization and molecular epidemiology of antimicrobial-resistant Pseudomonas aeruginosa in a veterinary teaching hospital. Bacterial specimens from surface and liquid samples (n = 165) located in five rooms were collected three times every 2 months, and antimicrobial susceptibility was subsequently determined by minimum inhibitory concentrations. The genomes of resistant strains were further analyzed using whole-genome sequencing. Among 19 P. aeruginosa isolates (11.5%, 19/165), sinks were the most frequent colonization site (53.3%), followed by rubber tubes (44.4%), and anesthesia-breathing circuit (33.3%). The highest resistance to gentamicin (47.4%), followed by piperacillin/tazobactam (36.8%), levofloxacin (36.8%), and ciprofloxacin (36.8%), was observed from 19 P. aeruginosa isolates, of which 10 were resistant strains. Of these 10 antimicrobial-resistant isolates, five were multidrug-resistant isolates, including carbapenem. From the multilocus sequence typing (MLST) analysis, five sequence types (STs), including a high-risk clone of human ST235 (n = 3), and ST244 (n = 3), ST606 (n = 2), ST485 (n = 1), and ST3405 (n = 1) were identified in resistant strains. Multiresistant genes were identified consistent with STs, except ST235. The MLST approach and single nucleotide polymorphism analysis revealed a link between resistant strains from ward rooms and those from examination, wound care, and operating rooms. The improvement of routine cleaning, especially of sink environments, and the continued monitoring of antimicrobial resistance of P. aeruginosa in veterinary hospitals are necessary to prevent the spread of resistant clones and ensure infection control.
Journal Title: Veterinary research communications
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!