Photo from academic.microsoft.com
Most 5-methylcytosine-binding domain proteins (such as MeCP2 and MBD1-6) contain a highly conserved 70 amino acid methyl-CpG-binding domain (MBD). The founding member of vertebrate MBD proteins, MBD2, is associated with… Click to show full abstract
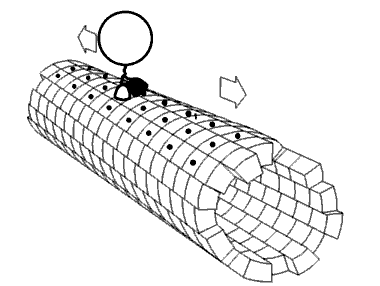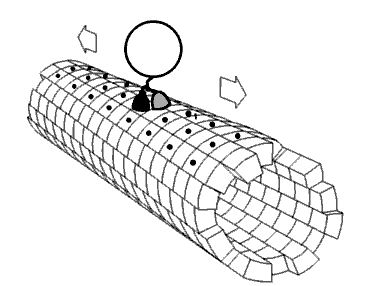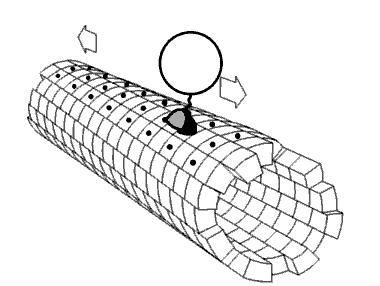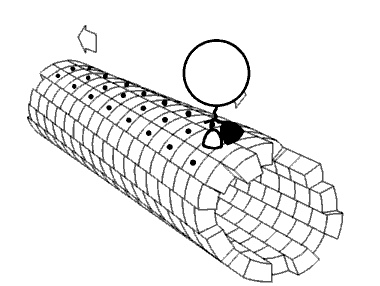
Most 5-methylcytosine-binding domain proteins (such as MeCP2 and MBD1-6) contain a highly conserved 70 amino acid methyl-CpG-binding domain (MBD). The founding member of vertebrate MBD proteins, MBD2, is associated with the Nucleosome Remodeling Deacetylase complex (NuRD) and exhibits DNA binding specificity for methylated CpG (mCpG) islands. MBD2 consists of the MBD, an intrinsically disorder region (IDR), and a coiled-coil region. The inclusion of the IDR in MBD2 increases binding affinity for mCpG. However, how MBD2 recognizes methylated DNA and the role of the IDR in influencing binding are not fully understood. To further elucidate the role of the IDR in DNA binding, we incorporated a series of biophysical techniques to examine movement of MBD2 without the IDR (MBD2MBD) or with a portion of the IDR (MBD2MBD+IDR). NMR and ZZ-exchange spectroscopy showed differences in the distribution and intramolecular exchange rate of the two protein constructs on 17bp dsDNA substrates that contain either CpG islands, mCpG sites, or no CpG sites. Single-molecule fluorescence tracking of quantum dot-labeled MBD2MBD and MBD2MBD+IDR on DNA tightropes containing CpG or mCpG reveals that both MBD2MBD and MBD2MBD+IDR search on DNA through 1D free diffusion and stably bind to methylated DNA. In conclusion, our data reveals how MBD2 dynamically achieves DNA binding specificity for methylated DNA and supports that the IDR of MBD2 modulates MBD2-binding dynamics. These results are important for further deciphering the molecular mechanisms of MBD and mCpG epigenetic regulation.
Journal Title: Biophysical Journal
Year Published: 2017
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!