Photo from wikipedia
SUMMARY High-dimensional data, such as those generated by single-cell RNA sequencing (scRNA-seq), present challenges in interpretation and visualization. Numerical and computational methods for dimensionality reduction allow for low-dimensional representation of… Click to show full abstract
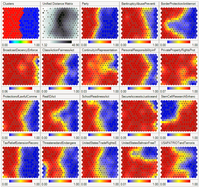
SUMMARY High-dimensional data, such as those generated by single-cell RNA sequencing (scRNA-seq), present challenges in interpretation and visualization. Numerical and computational methods for dimensionality reduction allow for low-dimensional representation of genome-scale expression data for downstream clustering, trajectory reconstruction, and biological interpretation. However, a comprehensive and quantitative evaluation of the performance of these techniques has not been established. We present an unbiased framework that defines metrics of global and local structure preservation in dimensionality reduction transformations. Using discrete and continuous real-world and synthetic scRNA-seq datasets, we show how input cell distribution and method parameters are largely determinant of global, local, and organizational data structure preservation by 11 common dimensionality reduction methods.
Journal Title: Cell reports
Year Published: 2020
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!