Photo from wikipedia
FGF/ERK signaling is crucial for the patterning and proliferation of cell lineages that comprise the mouse blastocyst. However, ERK signaling dynamics have never been directly visualized in live embryos. To… Click to show full abstract
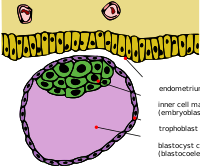
FGF/ERK signaling is crucial for the patterning and proliferation of cell lineages that comprise the mouse blastocyst. However, ERK signaling dynamics have never been directly visualized in live embryos. To address whether differential signaling is associated with particular cell fates and states, we generated a targeted mouse line expressing an ERK-kinase translocation reporter (KTR) that enables live quantification of ERK activity at single-cell resolution. 3D time-lapse imaging of this biosensor in embryos revealed spatially graded ERK activity in the trophectoderm prior to overt polar versus mural differentiation. Within the inner cell mass (ICM), all cells relayed FGF/ERK signals with varying durations and magnitude. Primitive endoderm cells displayed higher overall levels of ERK activity, while pluripotent epiblast cells exhibited lower basal activity with sporadic pulses. These results constitute a direct visualization of signaling events during mammalian pre-implantation development and reveal the existence of spatial and temporal lineage-specific dynamics.
Journal Title: Developmental cell
Year Published: 2020
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!