Photo from wikipedia
Macrolide antibiotics, such as erythromycin, bind to the nascent peptide exit tunnel (NPET) of the bacterial ribosome and modulate protein synthesis depending on the nascent peptide sequence. Whereas in vitro… Click to show full abstract
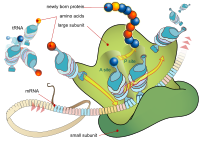
Macrolide antibiotics, such as erythromycin, bind to the nascent peptide exit tunnel (NPET) of the bacterial ribosome and modulate protein synthesis depending on the nascent peptide sequence. Whereas in vitro biochemical and structural methods have been instrumental in dissecting and explaining the molecular details of macrolide-induced peptidyl-tRNA drop-off and ribosome stalling, the dynamic effects of the drugs on ongoing protein synthesis inside live bacterial cells are far less explored. In the present study, we used single-particle tracking of dye-labeled tRNAs to study the kinetics of mRNA translation in the presence of erythromycin, directly inside live Escherichia coli cells. In erythromycin-treated cells, we find that the dwells of elongator tRNAPhe on ribosomes extend significantly, but they occur much more seldom. In contrast, the drug barely affects the ribosome binding events of the initiator tRNAfMet. By overexpressing specific short peptides, we further find context-specific ribosome binding dynamics of tRNAPhe, underscoring the complexity of erythromycin's effect on protein synthesis in bacterial cells.
Journal Title: Journal of molecular biology
Year Published: 2021
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!