Photo from wikipedia
The mythical Hydra was a beast quite adept at re-growing its heads...at least until it met Heracles. The much smaller and much more real lab animal doesn’t have heads per… Click to show full abstract
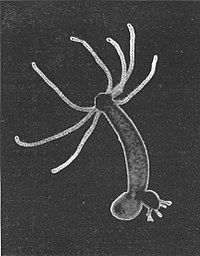
The mythical Hydra was a beast quite adept at re-growing its heads...at least until it met Heracles. The much smaller and much more real lab animal doesn’t have heads per se, but it shares its namesake’s ability to regenerate and replace tissue. Even in the absence of injury (heroic or otherwise), every cell in the animals’ body is replaced about every 20 days. Those cells differentiate from one of three lineages: endodermal epithelial, ectodermal endothelial, and interstitial. But how does a particular cell type, say a neuron or a stinging nematocyst, differentiate from its stem cell progenitor? It’s a beastly question that motivates research in many different animals, but one that a new resource from Celina Juliano’s lab at the University of California, Davis, might help those in the Hydra community slay. Juliano and her colleagues recently took on the labor of mapping cell differentiation trajectories for Hydra at single-cell resolution. They used a microfluidics-based singlecell sequencing technique called Drop-seq. The process is fairly straightforward, says Juliano: Hydra are dissociated into single cells—a mix of stem cells, differentiated cells, and those in transition—and an oil is used to create nanoliter-sized liquid droplets containing one cell and one barcoded ‘bead.’ A lysis buffer breaks open the cell to release the transcripts inside for sequencing; the barcoded beads meanwhile allow the researchers to keep track of the cellular origins of all that RNA. The first step of the analysis involved creating a digital expression matrix, with Hydra’s genes on one side and all the sequenced cell data on the other. From there, cells were grouped into clusters using the Seurat R package to construct a molecular map of cell states; another R package called URD, originally built by co-author Jeffrey Farrell at Harvard for zebrafish, was then used to build the different differentiation trajectories. Perhaps surprisingly, the team didn’t find any major surprises once they completed their initial map. “Hydra works as we think it works,” Juliano says. The resource however should provide a valuable starting point for now figuring out the molecular details that enable Hydra to maintain its homeostatic state. “We have this map of gene expression, and not just of stable cell types but expression as cells are starting to differentiate,” she says. “Now we can start to understand cell differentiation starting from a much more informed place.” Work with Hyra labors on.
Journal Title: Lab Animal
Year Published: 2019
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!