Photo from wikipedia
Significance Widespread biological communities are common, but little is known about how they assemble. A key question is how sets of trophically linked species (predators and their prey, hosts and… Click to show full abstract
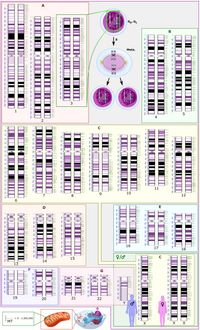
Significance Widespread biological communities are common, but little is known about how they assemble. A key question is how sets of trophically linked species (predators and their prey, hosts and parasites) spread to occupy current distributions. Do they disperse together, preserving ecological interactions, or separately, such that interactions are interrupted? This is central to assessing the potential for coevolution in a system and requires inference of species associations both over space and through time. Here, we use de novo genomic data and likelihood-based approaches to infer the assembly history of a multispecies community of Western Palearctic insect herbivores and parasitoid natural enemies—the two trophic groups that together comprise 50% of all animal species. How widespread ecological communities assemble remains a key question in ecology. Trophic interactions between widespread species may reflect a shared population history or ecological fitting of local pools of species with very different population histories. Which scenario applies is central to the stability of trophic associations and the potential for coevolution between species. Here we show how alternative community assembly hypotheses can be discriminated using whole-genome data for component species and provide a likelihood framework that overcomes current limitations in formal comparison of multispecies histories. We illustrate our approach by inferring the assembly history of a Western Palearctic community of insect herbivores and parasitoid natural enemies, trophic groups that together comprise 50% of terrestrial species. We reject models of codispersal from a shared origin and of delayed enemy pursuit of their herbivore hosts, arguing against herbivore attainment of “enemy-free space.” The community-wide distribution of species expansion times is also incompatible with a random, neutral model of assembly. Instead, we reveal a complex assembly history of single- and multispecies range expansions through the Pleistocene from different directions and over a range of timescales. Our results suggest substantial turnover in species associations and argue against tight coevolution in this system. The approach we illustrate is widely applicable to natural communities of nonmodel species and makes it possible to reveal the historical backdrop against which natural selection acts.
Journal Title: Proceedings of the National Academy of Sciences of the United States of America
Year Published: 2018
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!