Photo from wikipedia
The importance of adaptive immunity has been highlighted by the protection afforded by vaccination in the face of the current coronavirus pandemic. Much has been learned about the origins and… Click to show full abstract
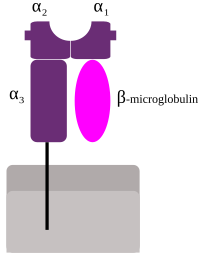
The importance of adaptive immunity has been highlighted by the protection afforded by vaccination in the face of the current coronavirus pandemic. Much has been learned about the origins and subsequent evolution of the antigen-specific receptors used by this crucial arm of the immune response. Among recent discoveries are the early appearance of three lymphocyte lineages in vertebrates with antigen-specific receptors based on leucine-rich repeats in jawless fish but on immunoglobulin (Ig) domains in jawed vertebrates (1), and the protoRAG transposon which created split genes that could recombine to generate diversity in the three antigen-specific receptors of jawed vertebrates (2). In contrast, there has been little agreement on the origin and subsequent evolution of cell surface molecules encoded by the major histocompatibility complex (MHC), which play central roles in adaptive immunity as the targets of T cell recognition. By discovering the W genes as a proposed intermediate in the evolution of MHC class I and class II genes, the paper by Okamura et al. (3) in PNAS provides a welcome advance. To appreciate this story, one must understand some aspects of the MHC molecules and the cells that recognize them (4). With well over 10,000 alleles among humans, the classical MHC molecules form the most polymorphic system currently known. Having been discovered as transplantation antigens, their true function is resistance to infectious pathogens and cancers. The high polymorphism is primarily due to a molecular arms race with pathogens, highlighting their importance in resistance to infectious disease. In addition, nonclassical class I molecules have evolved to carry out a wide variety of specialized functions. For example, natural killer (NK) cells of the innate immune system recognize certain classical and nonclassical class I molecules for both immune and nonimmune functions, the latter including placental blood supply for pregnancy (4, 5). Classical MHC molecules bind peptides within cells for presentation to T cells with T cell receptors (TCRs) composed of αand β-chains (4, 6). The αβ T cells bearing the coreceptor CD8 recognize classical class I molecules bound to peptides originating primarily in the cytoplasm and nucleus where viruses (and a few intracellular bacteria) replicate. CD8 αβ T cells are cytotoxic T lymphocytes which kill infected cells, preventing the release of new viruses. In contrast, the peptides presented by class II molecules originate largely from intracellular vesicles in contact with the extracellular space where most pathogens can be found, so responses by CD4 αβ T cells are more varied and nuanced, including crucial roles in regulation of most immune responses. Class I and class II molecules are built of similar protein domains but differ in the organization of these domains (Fig. 1), reflected in the intron–exon structure of their genes (7–11). Class II molecules are heterodimers of αand β-glycoproteins (encoded by A and B genes), each with a membrane-distal domain and membrane-proximal Ig-constant (Ig-C) domain, a transmembrane (TM) region, and a short cytoplasmic tail. In contrast, class I molecules are composed of one small Ig-C protein, β2-microglobulin (β2m), in noncovalent association with a large α (or heavy) glycoprotein chain with two membrane-distal domains followed by a membrane-proximal Ig-C domain, a TM region, and a cytoplasmic tail (4, 7–9). For both class I and class II molecules, the two membrane-distal domains together form a pair of broken α-helices atop a platform of β-strands (sometimes called an open-face sandwich or MHC fold) (8, 9). The groove between the α-helices and the β-sheet is where most of the polymorphic positions are found, in which each classical MHC allele binds a different set of peptides (4). The αβ TCRs of T cells and the killer-Ig receptors of human NK cells recognize the peptide and α-helices of the MHC molecules (4, 8, 9).
Journal Title: Proceedings of the National Academy of Sciences of the United States of America
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!