Photo from wikipedia
ABSTRACT Background: Homologous repair deficiency (HRD) causes double-strand break repair to be impeded, which is a common driver of carcinogenesis. However, the therapeutic and prognostic potential of HRD in invasive… Click to show full abstract
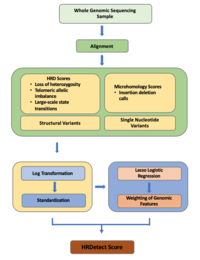
ABSTRACT Background: Homologous repair deficiency (HRD) causes double-strand break repair to be impeded, which is a common driver of carcinogenesis. However, the therapeutic and prognostic potential of HRD in invasive breast cancer (BRCA) has not been fully explored using comprehensive bioinformatics analysis. Materials and Methods: HRD score was defined as the unweighted sum of LOH, TAI, and LST scores and obtained from the previous study according to Theo A et al. To characterize BRCA tumor microenvironment (TME) subtypes, “ConsensusClusterPlus” R package was used to conduct unsupervised clustering. The xCell algorithm was utilized for tumor composition analysis to estimate the TME in TCGA-BRCA. A WGCNA analysis was conducted to uncover the gene coexpression modules and hub genes in the HRD-related gene module of BRCA. The functional enrichment study was carried out using Metascape. A novel analysis pipeline, Genetic Perturbation Similarity Analysis (GPSA), was used to explore the single-gene perturbation closely related to HRD based on 3048 stable knockdown/knockout cell lines. The prognostic variables were evaluated using univariate COX analysis. Kaplan-Meier (KM) survival analysis was performed to assess the prognostic potential of HRD score. Receiver operator characteristic (ROC) curve was utilized to judge the diagnostic utility. Drug sensitivity was estimated through the R package “oncoPredict” and Genomics of Drug Sensitivity in Cancer (GDSC) database. XSum algorithm was performed to screen the candidate small molecule drugs based on the connectivity map (CMAP) database. Results: Low HRD score suggested a better prognosis in BRCA patients. The tumor with low HRD score had considerably greater degree of infiltration of stromal cells and infiltration of immunocytes was significantly enhanced in the high HRD score group. Using WGCNA, ten co-expression modules were obtained. The turquoise module and 25 hub genes were identified as the most correlated with HRD in BRCA. Functional enrichment analysis revealed that the turquoise gene module was mainly concentrated in the “cell cycle” pathways. Candidate HRD-related gene signatures (MELK) were screened out through WGCNA and GPSA analysis pipeline and then validated on independent validation sets. A small molecule drug (Clofibrate) that has the potential to reverse the increase of high HRD score was screened out to improve oncological outcomes in BRCA. Molecular docking suggested MELK to be one of possible molecular targets in the Clofibrate treatment of BRCA. Conclusion: Based on bioinformatic analysis, we fully explored the therapeutic and prognostic potential of HRD in BRCA. A novel HRD-related gene signature (MELK) were identified through the combination of WGCNA and GPSA analysis. In addition, we detailed the TME landscape in BRCA and identified four unique TME subtypes in group with high or low HRD score group. Moreover, Clofibrate were screened out to improve oncological outcomes in BRCA by reversing the increase of high HRD score. Thus, our study contributes to the development of personalized clinical management and treatment regimens of BRCA.