Photo from wikipedia
Motivation: Comparing ribonucleic acid (RNA) secondary structures of arbitrary size uncovers structural patterns that can provide a better understanding of RNA functions. However, performing fast and accurate secondary structure comparisons… Click to show full abstract
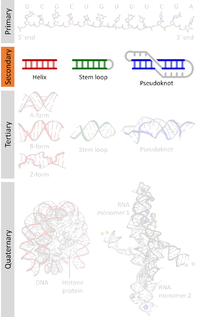
Motivation: Comparing ribonucleic acid (RNA) secondary structures of arbitrary size uncovers structural patterns that can provide a better understanding of RNA functions. However, performing fast and accurate secondary structure comparisons is challenging when we take into account the RNA configuration (i.e. linear or circular), the presence of pseudoknot and G‐quadruplex (G4) motifs and the increasing number of secondary structures generated by high‐throughput probing techniques. To address this challenge, we propose the super‐n‐motifs model based on a latent analysis of enhanced motifs comprising not only basic motifs but also adjacency relations. The super‐n‐motifs model computes a vector representation of secondary structures as linear combinations of these motifs. Results: We demonstrate the accuracy of our model for comparison of secondary structures from linear and circular RNA while also considering pseudoknot and G4 motifs. We show that the super‐n‐motifs representation effectively captures the most important structural features of secondary structures, as compared to other representations such as ordered tree, arc‐annotated and string representations. Finally, we demonstrate the time efficiency of our model, which is alignment free and capable of performing large‐scale comparisons of 10 000 secondary structures with an efficiency up to 4 orders of magnitude faster than existing approaches. Availability and Implementation: The super‐n‐motifs model was implemented in C ++. Source code and Linux binary are freely available at http://jpsglouzon.github.io/supernmotifs/. Contact: [email protected] Supplementary information: Supplementary data are available at Bioinformatics online.
Journal Title: Bioinformatics
Year Published: 2017
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!