Photo from wikipedia
Summary: Detecting and interpreting responsive modules from gene expression data by using network‐based approaches is a common but laborious task. It often requires the application of several computational methods implemented… Click to show full abstract
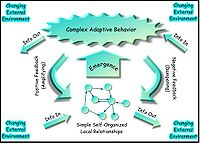
Summary: Detecting and interpreting responsive modules from gene expression data by using network‐based approaches is a common but laborious task. It often requires the application of several computational methods implemented in different software packages, forcing biologists to compile complex analytical pipelines. Here we introduce INfORM (Inference of NetwOrk Response Modules), an R shiny application that enables non‐expert users to detect, evaluate and select gene modules with high statistical and biological significance. INfORM is a comprehensive tool for the identification of biologically meaningful response modules from consensus gene networks inferred by using multiple algorithms. It is accessible through an intuitive graphical user interface allowing for a level of abstraction from the computational steps. Availability and implementation: INfORM is freely available for academic use at https://github.com/Greco‐Lab/INfORM. Supplementary information: Supplementary data are available at Bioinformatics online.
Journal Title: Bioinformatics
Year Published: 2018
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!