Photo from wikipedia
Antifungal susceptibility testing is an essential tool for guiding antifungal therapy. Reference methods are complex and usually only available in specialized laboratories. We have designed an expanded agar-based screening method… Click to show full abstract
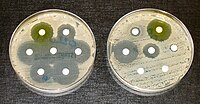
Antifungal susceptibility testing is an essential tool for guiding antifungal therapy. Reference methods are complex and usually only available in specialized laboratories. We have designed an expanded agar-based screening method for the detection of azole resistant Aspergillus fumigatus isolates. Normally, identification of resistance mechanisms is obtained only after sequencing the cyp51A gene and promoter. However, our screening method provides azole resistance detection and presumptive resistance mechanisms identification. A previous agar-based method consisting of four wells containing voriconazole, itraconazole, posaconazole and a growth control, detected azole resistance to clinical azoles. Here we have modified the concentrations of voriconazole and posaconazole to adapt to the updated EUCAST breakpoints against A. fumigatus. We have also, expanded the method to include environmental azoles to assess azole resistance and the azole resistance mechanism involved. We used a collection of A. fumigatus including 54 azole resistant isolates with Cyp51A modifications (G54, M220, G448S, TR53 , TR34 /L98H, TR46 /Y121F/T289A, TR34 /L98H/S297T/F495I), and 50 azole susceptible isolates with wild-type Cyp51A. The screening method detects azole-resistant A. fumigatus isolates when there is growth in any of the azole-containing wells after 48h. The growth pattern in the seven azoles tested helps determine the underlying azole resistance mechanism. This approach is designed for surveillance screening of A. fumigatus azole-resistant isolates and can be useful for the clinical management of patients prior to antifungal susceptibility testing confirmation.
Journal Title: Mycoses
Year Published: 2021
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!