Photo from wikipedia
For the study of upper gastrointestinal microbiota in humans, endoscopic sampling is the main source of information, which limits the understanding of healthy upper gastrointestinal microbiota. Rhesus monkeys show very… Click to show full abstract
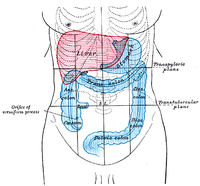
For the study of upper gastrointestinal microbiota in humans, endoscopic sampling is the main source of information, which limits the understanding of healthy upper gastrointestinal microbiota. Rhesus monkeys show very close similarity to humans in physiology, genetics, and behavior and act as the most suitable animal models for human diseases. ABSTRACT The strategy of adjusting the composition of gastrointestinal microbiota has shown great promise for the treatment of diseases. Currently, the relationship between gut microbes and human diseases is mainly presented by the fecal microbiota from the colon. Due to the limits of sampling, the healthy baseline of biogeographic microbiota in the human gastrointestinal tract remains blurry. Captive nonhuman primates (NHPs) present a “humanized” intestinal microbiome and may make up for the lack of atlas data for better understanding of the gut microbial composition and diseases. Therefore, the intestinal microbiota of 6 GIT regions of healthy rhesus monkeys were analyzed in this study; our results showed that Proteobacteria gradually decreased from the small intestine to the large intestine but Bacteroidetes gradually increased from the small intestine to the large intestine. Streptococcus and Lactobacillus can be used as markers to distinguish the small intestine from the large intestine. Sarcina is the most enriched in the middle site of the connection between the large intestine and the small intestine. Cyanobacteria are enriched in the small intestine, especially the duodenum and jejunum, and are absent in the large intestine. The lumenal microbiota of the small intestine is more susceptible to individual differences than is that of the large intestine. Metabolism and oxygen affect the distribution of the microbes, and the diversity of microbiota is the highest in the colon. Our results provide accurate comprehensive GIT microbiota data on nonhuman primates and will be beneficial for the better understanding of the composition of microbiota in the human gastrointestinal tract. IMPORTANCE For the study of upper gastrointestinal microbiota in humans, endoscopic sampling is the main source of information, which limits the understanding of healthy upper gastrointestinal microbiota. Rhesus monkeys show very close similarity to humans in physiology, genetics, and behavior and act as the most suitable animal models for human diseases. The present research made up for the lack of atlas data due to the ethical limitations of sampling in humans and provided baseline data on microbiota in 6 GIT regions of healthy NHPs. These important references will be beneficial for the better understanding of the regional organization and functions of gut microbial communities along the GIT and their relevance to conditions of health and disease.
Journal Title: Microbiology Spectrum
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!