Photo from wikipedia
Acinetobacter baumannii has emerged as a significant hospital pathogen, quickly becoming resistant to commonly prescribed antimicrobials. The present survey was done to evaluate the prevalence, antibiotic resistance pattern, and distribution… Click to show full abstract
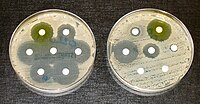
Acinetobacter baumannii has emerged as a significant hospital pathogen, quickly becoming resistant to commonly prescribed antimicrobials. The present survey was done to evaluate the prevalence, antibiotic resistance pattern, and distribution of antibiotic resistance genes amongst the A. baumannii strains isolated from fish, shrimp, and lobster samples. Four-hundred and fifty seafood samples (100 g each) were collected from Shiraz, Iran. Acinetobacter baumannii was determined using culture and biochemical tests. Pattern of antibiotic resistance and distribution of antibiotic resistance genes were determined using the disk diffusion and polymerase chain reaction, respectively. A. baumannii contamination rate amongst the examined seafood samples was 4.44%, with the higher contamination rate of fish samples (7.85%). A. baumannii isolates harbored the maximum resistance rate against tetracycline (85%), ampicillin (85%), gentamicin (70%), and erythromycin (60%). Resistance rates toward trimethoprim-sulfamethoxazole, ciprofloxacin, ceftazidime, and azithromycin were 55%, 45%, 35%, and 30%, respectively. The minimum rates of resistance were obtained against imipenem (10%) and chloramphenicol (15%). The most commonly detected antibiotic resistance genes were blaCITM (75%), blaSHV (70%), tetA (70%), qnrA (55%), blaVIM (50%), and aac(3)-IV (50%). aadA1, sul1, dfrA1, qnr, blaVIM, blaSIM, blaOXA-51, blaOXA-23, and blaOXA-58 genes were detected in 40%, 30%, 45%, 50%, 35%, 25%, 30%, and 20% of isolates, respectively. The role of seafood samples as a potential reservoirs of antibiotic-resistant A. baumannii strains was determined. However, further investigations are required to identify additional epidemiological features of A. baumannii in seafood samples.
Journal Title: Journal of Food Processing and Preservation
Year Published: 2023
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!