Photo from wikipedia
To characterize modulators of enzymatic activity, a continuous assay format will quantify with confidence the initial reaction velocity far better than an end point assay. When the progress curve is… Click to show full abstract
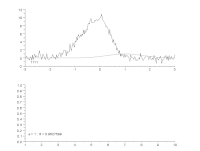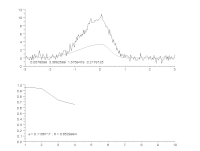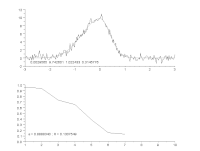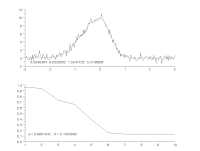
To characterize modulators of enzymatic activity, a continuous assay format will quantify with confidence the initial reaction velocity far better than an end point assay. When the progress curve is linear from the start and remains linear throughout the experiment, a single end point reading can approximate the reaction rate, but a kinetic assay will provide the rate more accurately. However, if there is a delay in the onset of the reaction, or there are other changes in reaction rate over time, then a single end point reading is insufficient. Thus, important information about the reaction can be missed or mischaracterized. While visual inspection is the most common way to determine the range of the progress curve from which to extract rate information, this method is not practical when thousands of progress curves are generated in an experimental day. We have developed an automated protocol to streamline and optimize the process. The heuristic algorithm analyzes the progress curve to identify different regions for characterization (e.g. lag, primary rate, secondary rate, final plateau) and then extract rates of interest for each of these regions. These rates are subsequently used to measure the specific activity of the enzyme under various assay conditions, and to characterize the modulation of that activity: simple dose-response testing, mode of inhibition analysis, time-dependent inhibition determination, reversibility testing, determination of residence time for reversible inhibitors, and kinact/KI analysis for irreversible inhibitors. We describe the algorithm and several applications as they apply to protein kinases using the PhosphoSens assay platform from AssayQuant within the Analyze module of the Scigilian data analysis package. Citation Format: Earl William May, Daniel Urul, Susan Cornell-Kennon, Zhibing Lu, Erik Schaefer, Sam Hoare, France Laliberté, Quay Vong, Paul Payette, Jean Marois. Automation of linear range determination: Enzymatic progress curve applications [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2023; Part 1 (Regular and Invited Abstracts); 2023 Apr 14-19; Orlando, FL. Philadelphia (PA): AACR; Cancer Res 2023;83(7_Suppl):Abstract nr 2766.
Journal Title: Cancer Research
Year Published: 2023
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!