Photo from wikipedia
Environmental DNA (eDNA) metabarcoding is an increasingly popular method for rapid biodiversity assessment. As with any ecological survey, false negatives can arise during sampling and, if unaccounted for, lead to… Click to show full abstract
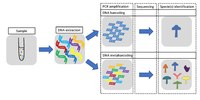
Environmental DNA (eDNA) metabarcoding is an increasingly popular method for rapid biodiversity assessment. As with any ecological survey, false negatives can arise during sampling and, if unaccounted for, lead to biased results and potentially misdiagnosed environmental assessments. We developed a multi-scale, multi-species occupancy model for the analysis of community biodiversity data resulting from eDNA metabarcoding; this model accounts for imperfect detection and additional sources of environmental and experimental variation. We present methods for model assessment and model comparison and demonstrate how these tools improve the inferential power of eDNA metabarcoding data using a case study in a coastal, marine environment. Using occupancy models to account for factors often overlooked in the analysis of eDNA metabarcoding data will dramatically improve ecological inference, sampling design, and methodologies, empowering practitioners with an approach to wield the high-resolution biodiversity data of next-generation sequencing platforms.
Journal Title: PLoS ONE
Year Published: 2020
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!