Photo from wikipedia
Systemic lupus erythematosus (SLE) is caused by a combination of genetic and environmental factors. Recently, analysis of a functional genome database of genetic polymorphisms and transcriptomic data from various immune… Click to show full abstract
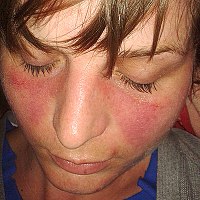
Systemic lupus erythematosus (SLE) is caused by a combination of genetic and environmental factors. Recently, analysis of a functional genome database of genetic polymorphisms and transcriptomic data from various immune cell subsets revealed the importance of the oxidative phosphorylation (OXPHOS) pathway in the pathogenesis of SLE. In particular, activation of the OXPHOS pathway is persistent in inactive SLE, and this activation is associated with organ damage. The finding that hydroxychloroquine (HCQ), which improves the prognosis of SLE, targets toll-like receptor (TLR) signaling upstream of OXPHOS suggests the clinical importance of this pathway. IRF5 and SLC15A4, which are regulated by polymorphisms associated with SLE susceptibility, are functionally associated with OXPHOS as well as blood interferon activity and metabolome. Future analyses of OXPHOS-associated disease-susceptibility polymorphisms, gene expression, and protein function may be useful for risk stratification of SLE.
Journal Title: Biomolecules
Year Published: 2023
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!