Photo from wikipedia
Existing tools to estimate cardiovascular (CV) risk have sub-optimal predictive capacities. In this setting, non-invasive imaging techniques and omics biomarkers could improve risk-prediction models for CV events. This study aimed… Click to show full abstract
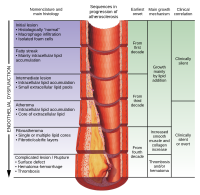
Existing tools to estimate cardiovascular (CV) risk have sub-optimal predictive capacities. In this setting, non-invasive imaging techniques and omics biomarkers could improve risk-prediction models for CV events. This study aimed to identify gene expression patterns in whole blood that could differentiate patients with severe coronary atherosclerosis from subjects with a complete absence of detectable coronary artery disease and to assess associations of gene expression patterns with plaque features in coronary CT angiography (CCTA). Patients undergoing CCTA for suspected coronary artery disease (CAD) were enrolled. Coronary stenosis was quantified and CCTA plaque features were assessed. The whole-blood transcriptome was analyzed with RNA sequencing. We detected highly significant differences in the circulating transcriptome between patients with high-degree coronary stenosis (≥70%) in the CCTA and subjects with an absence of coronary plaque. Notably, regression analysis revealed expression signatures associated with the Leaman score, the segment involved score, the segment stenosis score, and plaque volume with density <150 HU at CCTA. This pilot study shows that patients with significant coronary stenosis are characterized by whole-blood transcriptome profiles that may discriminate them from patients without CAD. Furthermore, our results suggest that whole-blood transcriptional profiles may predict plaque characteristics.
Journal Title: Biomedicines
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!