Photo from wikipedia
Improvements in microarray-based comparative genomic hybridization technology have allowed for high-resolution detection of genome wide copy number alterations, leading to a better definition of rearrangements and supporting the study of… Click to show full abstract
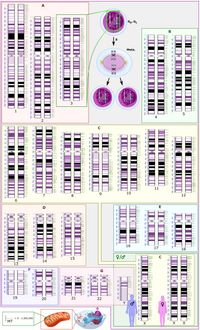
Improvements in microarray-based comparative genomic hybridization technology have allowed for high-resolution detection of genome wide copy number alterations, leading to a better definition of rearrangements and supporting the study of pathogenesis mechanisms. In this study, we focused our attention on chromosome 8p. We report 12 cases of 8p rearrangements, analyzed by molecular karyotype, evidencing a continuum of fragility that involves the entire short arm. The breakpoints seem more concentrated in three intervals: one at the telomeric end, the others at 8p23.1, close to the beta-defensin gene cluster and olfactory receptor low-copy repeats. Hypothetical mechanisms for all cases are described. Our data extend the cohort of published patients with 8p aberrations and highlight the need to pay special attention to these sequences due to the risk of formation of new chromosomal aberrations with pathological effects.
Journal Title: International Journal of Molecular Sciences
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!