Photo from wikipedia
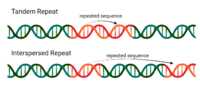
The multiple-locus variable-number analysis (MLVA) of tandem repeat (TR) for subtyping Listeria monocytogenes (LM) has proven to be reliable and fast. The study determined the MLVA genotypes of 60 isolates… Click to show full abstract
The multiple-locus variable-number analysis (MLVA) of tandem repeat (TR) for subtyping Listeria monocytogenes (LM) has proven to be reliable and fast. The study determined the MLVA genotypes of 60 isolates of L. monocytogenes recovered from cattle farms, abattoirs, and retail outlets in Gauteng province, South Africa. The distribution of the 60 L. monocytogenes isolates analyzed by type of sample was from raw beef (28, 46.7%), ready-to-eat (RTE), beef products (9, 15.0%), beef carcass swabs (9, 15.0%), cattle environment (6, 10.0%), and cattle faeces (8, 13.3%). The serogroups of the isolates were determined using polymerase chain reaction (PCR) and the MLVA genotypes based on six selected loci. The frequency of the serogroups detected 45.0% (27/60), 40.0% (24/60), 13.3% (8/60), and 1.7% (1/60) for serogroups 1/2a-3a (IIa), 4b-4d-4e (1Vb), 1/2c-3c (IIc), and 1/2b-3b (IIb), respectively. The MLVA successfully clustered the genetically related and differentiated non-related isolates, irrespective of their sources, sample types, and serogroups, as demonstrated by 16 MLVA pattern types detected. For serogroup 4b-4d-4e (IVb) there was no variation in tandem repeats LM-TR2, LM-TR4, and LM-TR6 which each contained only one allele, 02, 00, and 93 respectively. However, across the sources and sample types of isolates, there was variation in serogroup 4b-4d-4e (IVb) in tandem repeats LM-TR1, LM-TR3, and LM-TR5 which contained alleles (00, 03, 05), (14, 20, 22), and (14, 21, 25), respectively. Similar patterns of variation in the tandem repeat were detected in the other serogroups (1/2a-3a, 1/2b-3b, and 1/2c-3c). BioNumeric data analysis identified at least five types in Gauteng province. The MLVA epidemiologically clustered the related isolates and differentiated unrelated isolates.
Journal Title: Journal of food protection
Year Published: 2022
Link to full text (if available)
Share on Social Media: Sign Up to like & get
recommendations!