
Different types of disease‐causing noncoding variants revealed by genomic and gene expression analyses in families with X‐linked intellectual disability
Sign Up to like & getrecommendations! Published in 2021 at "Human Mutation"
DOI: 10.1002/humu.24207
Abstract: The pioneering discovery research of X‐linked intellectual disability (XLID) genes has benefitted thousands of individuals worldwide; however, approximately 30% of XLID families still remain unresolved. We postulated that noncoding variants that affect gene regulation or… read more here.
Keywords: noncoding variants; different types; intellectual disability; linked intellectual ... See more keywords
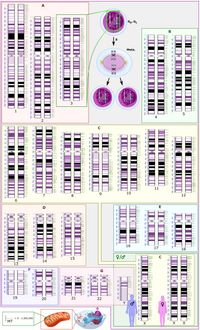
A method for scoring the cell type-specific impacts of noncoding variants in personal genomes
Sign Up to like & getrecommendations! Published in 2020 at "Proceedings of the National Academy of Sciences of the United States of America"
DOI: 10.1073/pnas.1922703117
Abstract: Significance Here we use the expression and accessibility data from a diverse set of cell types to learn a model for the dependence of the accessibility of a regulatory element on its DNA sequence and… read more here.
Keywords: noncoding variants; personal genome; accessibility; expression ... See more keywords

Prioritization and functional assessment of noncoding variants associated with complex diseases
Sign Up to like & getrecommendations! Published in 2018 at "Genome Medicine"
DOI: 10.1186/s13073-018-0565-y
Abstract: Unraveling functional noncoding variants associated with complex diseases is still a great challenge. We present a novel algorithm, Prioritization And Functional Assessment (PAFA), that prioritizes and assesses the functionality of genetic variants by introducing population… read more here.
Keywords: noncoding variants; associated complex; functional assessment; complex diseases ... See more keywords

Characterization of Coding/Noncoding Variants for SHROOM3 in Patients with CKD.
Sign Up to like & getrecommendations! Published in 2018 at "Journal of the American Society of Nephrology : JASN"
DOI: 10.1681/asn.2017080856
Abstract: Background Interpreting genetic variants is one of the greatest challenges impeding analysis of rapidly increasing volumes of genomic data from patients. For example, SHROOM3 is an associated risk gene for CKD, yet causative mechanism(s) of… read more here.
Keywords: noncoding variants; variants shroom3; characterization coding; ckd ... See more keywords